Epoxy Surfaces
for coupling via the N-terminus of biochemical species
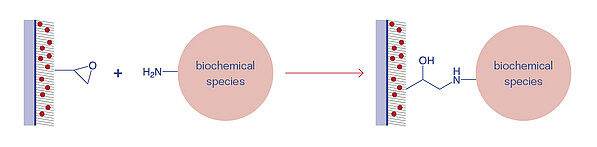
Epoxides are cyclic ethers with a highly strained three ring that can easily react with nucleophilic groups, e.g. amines, thiols, and hydroxyl groups. The Epoxy surface is uncharged and temperature-stable up to 40°C. Compared to NHS or PDITC, the Epoxy surface is more stable against humidity, and thus, has a longer shelf-life.
PolyAn equips glass slides, coverslips, polymer slides and 96-well plates with reactive surfaces. Please do not hesitate to contact us, if you would like to functionalize a different format or substrate with our 2D-Epoxy or 3D-Epoxy surface.
Selected Publications
Epoxy surface for the immobilization of Viruses:
- Broich, L. et al., `Single influenza A viruses induce nanoscale cellular reprogramming at the virus-cell interface´, Nature Commun., 2025, 16, 3846. DOI: 10.1038/s41467-025-58935-8.
- Osman, M.K. et al., `The bat influenza A virus subtype H18N11 induces nanoscale MHCII clustering upon host cell attachment´, Nature Commun., 2025, 16, 3847. DOI: 10.1038/s41467-025-58834-y.
Epoxy surface for the immobilization of Peptides:
- Shen, X. et al., `A Pentavalent HIV-1 Subtype C Vaccine Containing Computationally Selected gp120 Strains Improves the Breadth of V1V2 Region Responses´, Vaccines, 2025, 13, 133. DOI: 10.3390/vaccines13020133.
- Vanshylla, K. et al., `Mosaic HIV-1 vaccine and SHIV challenge strain V2 loop sequence identity and protection in primates´, npj Vaccines, 2024, 9, 179. DOI: 10.1038/s41541-024-00974-1.
- Cai, F. et al., `Structural and genetic convergence of HIV-1 neutralizing antibodies in vaccinated nonhuman primates´, PLoS Pathogens, 2021, 17, 1009624. DOI: 10.1371/journal.ppat.1009624.
- Saunders, K.O. et al., `Lipid nanoparticle encapsulated nucleoside-modified mRNA vaccines elicit polyfunctional HIV-1 antibodies comparable to proteins in nonhuman primates´, npj Vaccines, 2021, 6, 50. DOI: 10.1038/s41541-021-00307-6.
- Gorini, G.et al., `Engagement of monocytes, NK cells, and CD4+ Th1 cells by ALVAC-SIV vaccination results in a decreased risk of SIVmac251 vaginal acquisition´, PLoS Pathogens, 2020, 16, 1008377. DOI: 10.1371/journal.ppat.1008377.
- Schneider, J.R: et al., `A MUC16 IgG Binding Activity Selects for a Restricted Subset of IgG Enriched for Certain Simian Immunodeficiency Virus Epitope Specificities´, J. Virol., 2020, 94, 01246-19. DOI: 10.1128/JVI.01246-19.
- Shen, X. et al., `HIV-1 Vaccine Sequences Impact V1V2 Antibody Responses: A Comparison of Two Poxvirus Prime gp120 Boost Vaccine Regimens´, Sci. Rep., 2020, 10, 2093. DOI: 10.1038/s41598-020-57491-z.
- Dennis, M. et al., `Coadministration of CH31 Broadly Neutralizing Antibody Does Not Affect Development of Vaccine-Induced Anti-HIV-1 Envelope Antibody Responses in Infant Rhesus Macaques´, J. Virol., 2019, 93, 01783-18. DOI: 10.1128/JVI.01783-18.
- Han, Q. et al., `Difficult-to-neutralize global HIV-1 isolates are neutralized by antibodies targeting open envelope conformations´, Nature Commun., 2019, 10, 2898. DOI: 10.1038/s41467-019-10899-2.
- Jones, A.T. et al., `HIV-1 vaccination by needle-free oral injection induces strong mucosal immunity and protects against SHIV challenge´, Nature Commun., 2019, 10, 798. DOI: 10.1038/s41467-019-08739-4.
- Nelson, A.N. et al., `Simian-Human Immunodeficiency Virus SHIV.CH505-Infected Infant and Adult Rhesus Macaques Exhibit Similar Env-Specific Antibody Kinetics, despite Distinct T-Follicular Helper and Germinal Center B Cell Landscapes´, J. Virol., 2019, 93, 00168-19. DOI: 10.1128/JVI.00168-19.
- Sande, C.J. et al., `Comprehensive profiling of antibodies against multiple infectious diseases in serum and the airway mucosa using synthetic peptide-based linear epitope microarrays´, bioRxiv, 2018, ---, ---. DOI: 10.1101/462689 .
- Schiffner, T. et al., `Structural and immunologic correlates of chemically stabilized HIV-1 envelope glycoproteins´, PLoS Pathogens, 2018, 14, 1006986. DOI: 10.1371/journal.ppat.1006986.
- Wen, Y. et al., `Generation and characterization of a bivalent protein boost for future clinical trials: HIV-1 subtypes CR01_AE and B gp120 antigens with a potent adjuvant´, PLOS one, 2018, 13, 194266. DOI: 10.1371/journal.pone.0194266.
- Phillips, B. et al., `Impact of Poxvirus Vector Priming, Protein Coadministration, and Vaccine Intervals on HIV gp120 Vaccine-Elicited Antibody Magnitude and Function in Infant Macaques´, Clin. Vacc. Immun., 2017, 24, 00231-17. DOI: 10.1128/CVI.00231-17.
- Shen, X. et al., `HIV-1 gp120 and Modified Vaccinia Virus Ankara (MVA) gp140 Boost Immunogens Increase Immunogenicity of a DNA/MVA HIV-1 Vaccine´, J. Virol., 2017, 91, 01077-17. DOI: 10.1128/JVI.01077-17.
- Shen, X. et al., `Cross-Linking of a CD4-Mimetic Miniprotein with HIV-1 Env gp140 Alters Kinetics and Specificities of Antibody Responses against HIV-1 Env in Macaques´, J. Virol., 2017, 91, 00401-17. DOI: 10.1128/JVI.00401-17.
- Zurawski, G. et al., `Superiority in Rhesus Macaques of Targeting HIV-1 Env gp140 to CD40 versus LOX-1 in Combination with Replication-Competent NYVAC-KC for Induction of Env-Specific Antibody and T Cell Responses´, J. Virol., 2017, 91, 01596-16. DOI: 10.1128/JVI.01596-16.
- Rohe, A. et al., `Identification of peptidic substrates for the human kinase Myt1 using peptide microarrays´, Bioorg. Med. Chem., 2015, 23, 4936. DOI: 10.1016/j.bmc.2015.05.021.
- Stephenson, K.E. et al., `Quantification of the epitope diversity of HIV-1-specific binding antibodies by peptide microarrays for global HIV-1 vaccine development´, J. Immunol. Meth., 2015, 416, 105. DOI: 10.1016/j.jim.2014.11.006.
- Schönberg, A. et al., `The Peptide Microarray ‘‘ChloroPhos1.0’’ Identifies New Phosphorylation Targets of Plastid Casein Kinase II (pCKII) in Arabidopsis thaliana´, PLoS One, 2014, 9, 108344. DOI: 10.1371/journal.pone.0108344.
- Rapsch, K. et al., `Identification of antimicrobial peptides and immobilization strategy suitable for a covalent surface coating with biocompatible properties´, Bioconjugate Chem., 2014, 25, 308. DOI: 10.1021/bc4004469.
- Rauh, D. et al., `An acetylome peptide microarray reveals specificities and deacetylation substrates for all human sirtuin isoforms´, Nature Commun., 2013, 4, 2327. DOI: 10.1038/ncomms3327.
Epoxy surface for the immobilization of Oligonucleotides (DNA/RNA):
- Warmt, C. et al., `An experimental comparison between primer and nucleotide labelling to produce RPA-amplicons used for multiplex detection of antibiotic resistance genes´, Sci. Rep., 2023, 13, 15734. DOI: 10.1038/s41598-023-42830-7.
- Warmt, C. et al., `Investigation and validation of labelling loop mediated isothermal amplification (LAMP) products with different nucleotide modifications for various downstream analysis´, Sci. Rep., 2022, 12, 7137. DOI: 10.1038/s41598-022-11320-7.
- Warmt, C. et al., `Using Cy5‑dUTP labelling of RPA‑amplicons with downstream microarray analysis for the detection of antibiotic resistance genes´, Sci. Rep., 2021, 11, 20137. DOI: 10.1038/s41598-021-99774-z.
- Díaz-Betancor, Z. et al., `Photoclick chemistry to create dextran-based nucleic acid microarrays´, Anal. Bioanal. Chem., 2019, 411, 6745. DOI: 10.1007/s00216-019-02050-3.
- Sikora, K. et al., `A Universal Microarray Detection Method for Identification of Multiple Phytophthora spp. Using Padlock Probes´, Amer. Phytopath. Soc., 2012, 102, 635. DOI: 10.1094/PHYTO-11-11-0309.
Epoxy surface for the immobilization of Proteins and Antibodies:
- Rudokas, V. et al., `Novel monoclonal antibodies against house dust mite allergen Der p 21 and their application to analyze allergen extracts´, PeerJ, 2024, 12, 17233. DOI: 10.7717/peerj.17233.
- Röckendorf, N. et al., `Parallel detection of multiple biomarkers in a point‑of‑care‑competent device for the prediction of exacerbations in chronic inflammatory lung disease´, Sci. Rep., 2024, 14, 12830. DOI: 10.1038/s41598-024-62784-8.
- Silimavicius, L. et al., `Microarray-based evaluation of selected recombinant timothy grass allergens expressed in E. Coli and N. Benthamiana´, BMC Biotechnol., 2024, 24, 72. DOI: 10.1186/s12896-024-00902-0.
- DiNatale, C. et al., `Highly sensitive detection of the neurodegenerative biomarker Tau by using the concentration effect of the pyro-electrohydrodynamic jetting´, Biosens. Bioelectron., 2023, 254, 116234. DOI: 10.1016/j.bios.2024.116234.
- DiNatale, C. et al., `Optimization of Chemical Protein Conjugation on Activated Glass Surfaces for the Development of an Innovative Biosensor for Testing Astronaut Health Biomarkers at Picogram Levels During Spaceflight´, SSRN, 2023, DOI: 10.2139/ssrn.4479678 .
- Allelein, S. et al., `Prostate-Specific Membrane Antigen (PSMA)-Positive Extracellular Vesicles in Urine: A Potential Liquid Biopsy Strategy for Prostate Cancer Diagnosis?´, Cancers, 2022, 14, 2987. DOI: 10.3390/cancers14122987.
- Allelein, S. et al., `Potential and challenges of specifically isolating extracellular vesicles from heterogeneous populations´, Sci. Rep., 2021, 11, 11585. DOI: 10.1038/s41598-021-91129-.
- Hettegger, P. et al., `High similarity of IgG antibody profiles in blood and saliva opens opportunities for saliva based serology´, PLoS One, 2019, 14, 218456. DOI: 10.1371/journal.pone.0218456.
- Peter, H. et al., `Lab-on-a-Chip Device for Rapid Measurement of Vitamin D Levels´, Meth. Mol. Biol., 2018, 35, 477. DOI: 10.1007/978-1-4939-7614-0_35.
- Peter, H. et al., `Lab-on-a-Chip Proteomic Assays for Psychiatric Disorders´, Adv. Exp. Med. Biol., 2017, 33, 339. DOI: 10.1007/978-3-319-52479-5_33.
- Soria, J. et al., `Tear proteome analysis in ocular surface diseases using label-free LC-MS/MS and multiplexed microarray biomarker validation´, Sci. Rep., 2017, 7, 17478. DOI: 10.1038/s41598-017-17536-2.
- Gerdtsson, A.S. et al., `Evaluation of Solid Supports for Slide- and Well-Based Recombinant Antibody Microarrays´, Microarrays, 2016, 5, 16. DOI: 10.3390/microarrays5020016.
Epoxy surface for the immobilization of Glycans:
- Kooner, A.S. et al., `Broad-Spectrum Legionaminic Acid-Specific Antibodies in Pooled Human IgGs Revealed by Glycan Microarrays with Chemoenzymatically Synthesized Nonulosonosides´, Molecules, 2024, 29, 3980. DOI: 10.3390/molecules29163980.
- Parhi, L. et al., `Placental colonization by Fusobacterium nucleatum is mediated by binding of the Fap2 lectin to placentally displayed Gal-GalNAc´, Cell Reports, 2022, 38, 110537. DOI: 10.1016/j.celrep.2022.110537.
- Noy-Porat, T. et al., `Therapeutic antibodies, targeting the SARS-CoV-2 spike N-terminal domain, protect lethally infected K18-hACE2 mice´, iScience, 2021, 24, 102479. DOI: 10.1016/j.isci.2021.102479.
- Shanthamurthy, C.D. et al., `Heparan Sulfate Mimetics Differentially Affect Homologous Chemokines and Attenuate Cancer Development´, J. Med. Chem., 2021, 64, 3367. DOI: 10.1021/acs.jmedchem.0c01800.
- Bashir, S. et al., `Association between Neu5Gc carbohydrate and serum antibodies against it provides the molecular link to cancer: French NutriNet-Santé study´, BMC Medicine, 2020, 18, 262. DOI: 10.1186/s12916-020-01721-8.
- Shanthamurthy, C.D. et al., `ABO Antigens Active Tri- and Disaccharides Microarray to Evaluate C-type Lectin Receptor Binding Preferences´, Sci. Rep., 2018, 8, 6603. DOI: 10.1038/s41598-018-24333-y.
- Ben-Arye, S.L. et al., `Profiling Anti-Neu5Gc IgG in Human Sera with a Sialoglycan Microarray Assay´, J. Vis. Exp., 2017, 125, 56094. DOI: 10.3791/56094.
