Microarray Slides for DNA applications
DNA Microarrays are two-dimensional arrays of DNA capture strands printed onto a solid substrate used to assay a large number of DNA variants within a sample for multiplexed high-throughput screening.
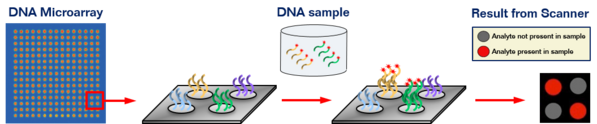
All of PolyAn’s reactive surfaces are completely transparent. They are characterized by a low lot-to-lot variation that is specified and monitored by using contact angle measurements as well as qualitative test methods.
A flyer showing the most commonly used functionalized surfaces for DNA Microarrays available from PolyAn can be downloaded here.
More hydrophobic surfaces may result in reduced spot diameter, depending on the spotting buffer composition. Results may vary based on buffers, sample preparation, spotting and scanning instruments.
Please note, that in order to facilitate handling of the glass slides PolyAn also offers a range of useful accessories and reagents.
Selected Publications
- Botti, V. et al., `Interaction between miR4749 and Human Serum Albumin as Revealed by Fluorescence, FRET, Atomic Force Spectroscopy and Computational Modelling´, Int. J. Mol. Sci. 2022, 23, 1291. DOI: 10.3390/ijms23031291.
- Warmt, C. et al., `Investigation and validation of labelling loop mediated isothermal amplification (LAMP) products with different nucleotide modifications for various downstream analysis´, Sci. Reports 2022, 12, 7137. DOI: 10.1038/s41598-022-11320-7.
- Karadimas, D. et al., `LATE-PCR for LoC Molecular Diagnostics Devices and Ist Application to the Sensitive Detection of SARS-CoV-2´, Eng. Proc. 2021, 6, 43. DOI: 10.3390/I3S2021Dresden-10076.
- Wang, Z. et al., `Detection of genetic variation and base modifications at base-pair resolution on both DANN and RNA´, Commun. Biology 2021, 4, 128. DOI: 10.1038/s42003-021-01648-7.
- Warmt, C. et al., `Using Cy5‑dUTP labelling of RPA‑amplicons with downstream microarray analysis for the detection of antibiotic resistance genes´, Sci. Reports 2021, 11, 20137. DOI: 10.1038/s41598-021-99774-z.
- Wolff, N. et al., `Full pathogen characterisation: species identification including the detection of virulence factors and antibiotic resistance genes via multiplex DNA‑assays´, Sci. Reports 2021, 11, 6001. DOI: 10.1038/s41598-021-85438-5.
- Díaz-Betancor, Z. et al., `Photoclick chemistry to create dextran-based nucleic acid microarrays´, Anal. Bioanal. Chem. 2019, 411, 6745. DOI: 10.1007/s00216-019-02050-3.
- Kurth, T. et al., `Development of Aptamer-Based TID Assays Using Thermophoresis and Microarrays´, Biosensors 2019, 9, 124. DOI: 10.3390/bios9040124.
- Prante, M. et al., `Characterization of an Aptamer Directed against 25-Hydroxyvitamin D for the Development of a Competitive Aptamer-Based Assay´, Biosensors 2019, 9, 134. DOI: 10.3390/bios9040134.
- Heilkenbrinker, A. et al., `Identification of the Target Binding Site of […] Aptamers and Its Exploitation for […] Detection´, Anal. Chem. 2015, 87, 677. DOI: 10.1021/ac5034819.
- Sikora, K. et al., `A Universal Microarray Detection Method for Identification of Multiple Phytophthora spp. Using Padlock Probes´, Amer. Phytopath. Soc. 2012, 102, 635. DOI: 10.1094/PHYTO-11-11-0309.
