Dual-color encoded Multiplex Beads
PolyAn's dual-color encoded beads have been developed specifically for read-out using a fluorescence microscope in combination with a suitable pattern recognition software. This encoding principle is illustrated in the image below.
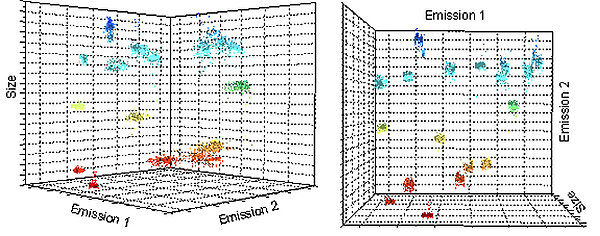
Dual-color encoded beads allow a narrower definition of the bead population, but have higher requirements regarding the read-out software. Using size as well as fluorescence makes it easier to differentiate between populations. The overall system becomes more robust and the requirements with regards to the read-out system can be reduced.
PolyAn Dual-color Multiplex Beads
The standard packaging volume is 1.5 mL/population with a solids content of 0.5%.
Dye-encoding for Multiplexing
PolyAn offers both dual-color encoding and single-dye-encoding. The spectral characteristics of the dyes used for the dual-color encoded system are:
| Color 1: | Excitation 420 - 480 nm | Color 2: | Excitation 515 - 540 nm |
| Emission 485 - 540 nm | Emission 535 - 570 nm |
The definition and differentiation between bead populations used in multiplex bead assays can be achieved using a combination of fluorescence emission wavelength (color), fluorescence intensity, and bead size. By combining fluorescence encoding and different particle size populations up to 100 populations can be defined.
Please do not hesitate to contact us, if you are interested in a set of multiplex bead populations that is tailored to your specific application and/or read-out system.
Selected publications
- Atiyas, Y. et al., `Combining time domain modulation optofluidics and high dynamic range imaging for multiplexed, high throughput digital droplet assays´, Microsys. Nanoeng., 2025, 11, 93. DOI: 10.1038/s41378-025-00918-2.
- Geithe, C. et al., `A multiplex microchamber diffusion assay for the antibody-based detection of microRNAs on randomly ordered microbeads´, Biosens. Bioelectron., 2024, 18, 100484. DOI: 10.1016/j.biosx.2024.100484.
- Dinter, F. et al., `Immobilisation of Lipophilic and Amphiphilic Biomarker on Hydrophobic Microbeads´, bioRxiv, 2023, DOI: 10.1101/2023.01.10.523433.
- Schmidt, C. et al., `A multiparametric fluorescence assay for screening aptamer–protein interactions based on microbeads´, Sci. Rep., 2022, 12, 2961. DOI: 10.1038/s41598-022-06817-0.
- Nawaz, S. et al., `Rapid Detection of Biofilm Formation by Zoonotic Serovars of Salmonella enterica and Avian Pathogenic E. coli Isolates from Poultry´, Pak. Vet. J., 2020, 40, 527. DOI: 10.29261/pakvetj/2020.066.
- Schmidt, C. et al., `Streptavidin Homologues for Applications on Solid Surfaces at High Temperatures´, Langmuir, 2020, 36, 628. DOI: 10.1021/acs.langmuir.9b02339.
- Choi, Y. et al., `A new reporter design based on DNA origami nanostructures for quantification of short oligonucleotides using microbeads´, Sci. Rep., 2019, 9, 4769. DOI: 10.1038/s41598-019-41136-x.
- Dinter, F. et al., `Simultaneous detection and quantification of DNA and protein biomarkers in spectrum of cardiovascular diseases in a microfluidic microbead chip´, Anal. Bioanal. Chem., 2019, 411, 7725. DOI: 10.1007/s00216-019-02199-x.
- Herrmann, A. et al., `Spatial Separation of Microbeads into Detection Levels by a Bioorthogonal Porous Hydrogel for Size-Selective Analysis and Increased Multiplexity´, Anal. Chem., 2019, 91, 8484. DOI: 10.1021/acs.analchem.9b01586.
- Olowe, O.A. et al., `Phylogenetic grouping and biofilm formation of multidrug resistant Escherichia coli isolates from humans, animals and food products in South-West Nigeria´, Sci. African, 2019, 6, 158. DOI: 10.1016/j.sciaf.2019.e00158.
- Liebsch, C. et al., `Solid-phase microbead array for multiplex O-serotyping of Escherichia coli´, Microchim. Acta, 2017, 184, 1405. DOI: 10.1007/s00604-017-2088-4.
- Schiebel, J. et al., `Genotypic and Phenotypic Characteristics Associated with Biofilm Formation by Human Clinical Escherichia coli Isolates of Different Pathotypes´, Appl. Environ. Microbiol., 2017, 83, 166017. DOI: 10.1128/AEM.01660-17.
- Schmidt, C. et al., `Multiplex localization of sequential peptide epitopes by use of a planar microbead chip´, Anal. Chim. Acta, 2016, 908, 150. DOI: 10.1016/j.aca.2015.12.030.
- Rödiger, S. et al., `Nucleic acid detection based on the use of microbeads: a review´, Microchim. Acta, 2014, 181, 1151. DOI: 10.1007/s00604-014-1243-4.
- Frömmel, U. et al., `Adhesion of Human and Animal Escherichia coli Strains in Association with Their Virulence-Associated Genes and Phylogenetic Origins´, Appl. Environ. Microbiol., 2013, 79, 5814. DOI: 10.1128/AEM.01384-13.
- Rödiger, S. et al., `A Highly Versatile Microscope Imaging Technology Platform for the Multiplex Real-Time Detection of Biomolecules and Autoimmune Antibodies´, Adv. Biochem. Eng. Biotechnol., 2013, 133, 35. DOI: 10.1007/10_2011_132.
- Rödiger, S. et al., `Mikropartikelsysteme für die Nukleinsäurediagnostik´, BioSpectrum, 2013, 19, 153. DOI: 10.1007/s12268-013-0287.
- Frömmel, U. et al., `Multiplex-PCR-Mikropartikel-Assay zum Nachweis bakterieller Gene´, Multiparameteranalytik in Forschung und Praxis, 2011, ISBN: 978-3-89967-703-4. DOI: 10.13140/2.1.4032.6084.
- Grossmann, K. et al., `Multiplex Assessment of Non-Organ-Specific Autoantibodies with a Novel Microbead-Based Immunoassay´, Cytometry A, 2011, 79, 118. DOI: 10.1002/cyto.a.21009.
